BINDER
 On Identification and Retrieval of Near-Duplicate Biological Images: a New Dataset and Protocol
On Identification and Retrieval of Near-Duplicate Biological Images: a New Dataset and Protocol
T.E. Koker, S.S. Chintapalli, S. Wang, B.A. Talbot, D. Wainstock, M. Cicconet, and M. Walsh
Paper: binder.pdf
Dataset: https://doi.org/10.7910/DVN/9YTD9Z
Code: https://github.com/hms-idac/BINDER
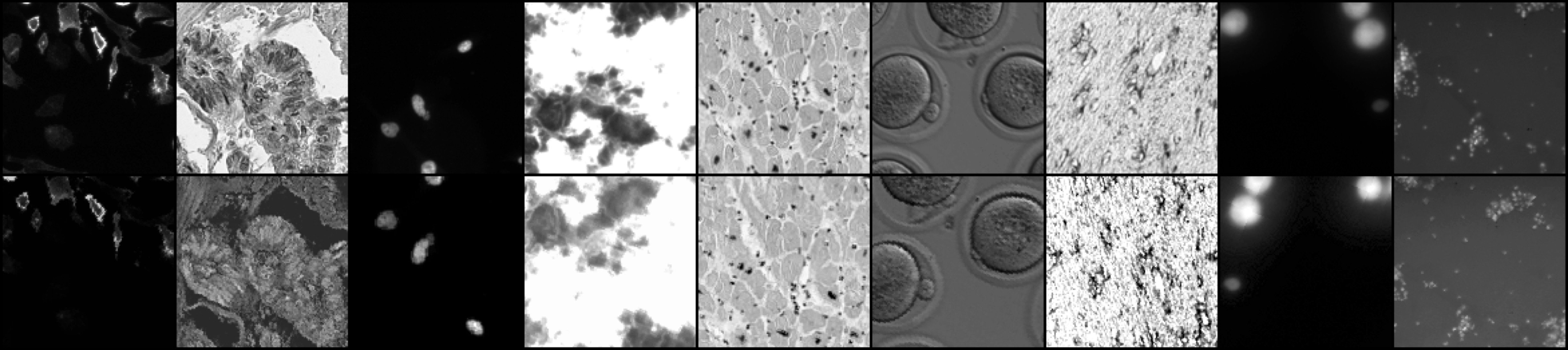
Abstract: Manipulation and re-use of images in scientific pub- lications is a growing issue, not only for biomedical publishers, but also for the research community in general. In this work we introduce BINDER – Bio-Image Near-Duplicate Examples Repository, a novel dataset to help researchers develop, train, and test models to detect same-source biomedical images. BINDER contains 7,490 unique image patches – the training set – as well as 1,821 patches – split in validation and test sets – with accompanying manipulations obtained by means of transforma- tions including rotation, translation, scale, perspective transform, contrast adjustment and/or compression artifacts. In addition, we show how novel adaptations of existing image retrieval and metric learning models, when trained on this dataset, can be applied to achieve high-accuracy inference results, creating a baseline for future work. In aggregate, we thus present a supervised protocol for near-duplicate image identification and retrieval without any “real-world” training example.
This is part of the Image Forensics project: https://hms-idac.github.io/ImageForensics/